Cladistics Definition
Cladistics refers to a biological classification system that involves the categorization of organisms based on shared traits. Organisms are typically grouped by how closely related they are and thus, cladistics can be used to trace ancestry back to shared common ancestors and the evolution of various characteristics. Although the classification of organisms began in the early 1900’s, cladistic analysis and specific methodology first originated in the 1960’s by Willi Hennig, referred to as “phylogenetic systematics”. This process involves creating phylogenies using morphological and molecular data to visualize evolutionary history and relationships between species.
Cladistic Methodologies
Cladistic methodologies involve the application of various molecular, anatomical, and genetic traits of organisms. Therefore, with the advent of computational modelling and molecular techniques (e.g., polymerase chain reaction [PCR]) cladistics are often used in evolutionary biology for the construction of phylogenetic trees. Cladistic data is also used to create cladograms (shown below), which consist of diagrams proposing a hypothesis of phylogenetic relationships between species based on shared characteristics. Thus, depending on a given dataset, the resulting cladogram may differ. For example, a cladogram based purely on morphological traits may produce different results from one constructed using genetic data. Today, highly advanced computational methods permit the use of multiple datasets to construct more accurate cladograms. Careful scientific analysis is required to rationally determine which cladogram may be a more correct representation than others.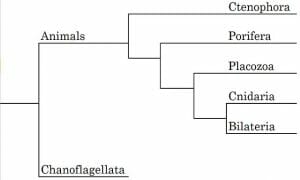
Cladistic Character States
In the field of cladistics, specific terminology is used to describe particular characteristics, termed “character states” among groups of organisms. The following are common terms used to describe such character states (illustrated below):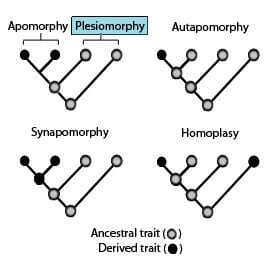
Plesiomorphy
Plesiomorphy refers to the ancestral traits that a taxon retains throughout evolution. Two or more taxa can share plesiomorphies but reside in different groups. When this occurs, this is termed “symplesiomorphies”. An example of a symplesiomorphy is quadrupedalism, or the ability to walk on four legs. Since this is an ancestral trait exhibited by reptiles, amphibians, and other taxa, this is a symplesiomorphy for mammals. Therefore, while symplesiomorphies can show distant evolutionary trends, it cannot be used to demonstrate more recently evolved characteristics.
Apomorphy
Apomorphy refers to a derived state used to define specific clades. Apomorphy can be further subdivided into “autapomorphies” and “synapomorphies”. Autapomorphies refer to traits that are exhibited only by one species or group, whereas synapomorphies refer to entire clades which can be classified by the presence of a particular trait. An example of synapomophy includes the presence of digits, shared by all tetrapods. An example of an autapomorphy is the capacity of human verbal speech, which is not exhibited by other primates, and is thus, a distinguishing human trait.
Homoplasy
Homoplasy refers to a character state that is shared by at least two organisms but is not found in the common ancestor or predecessor. Thus, the trait is aid to have evolved as a result of convergence or a reversal. A famous example of homoplasy is the evolution of warm-bloodedness in both mammals and birds, despite the absence of this trait from the common ancestor. Therefore, evidence indicates that the trait of warm-bloodedness must have evolved separately within each clade.
Quiz
1. Mammals and birds are both warm-blooded and share a common tetrapod ancestor. This is an example of:
A. Plesiomorphy
B. Autapomorphy
C. Apomorphy
D. Homoplasy
2. The evolution of bipedalism in humans compared to other primates is an example of:
A. Homoplasy
B. Autapomorphy
C. Synapomorphy
D. Synplesomorphy
References
- Bardin J, Rouget I, Yacobucci MM, Cecca F. (2014). Increasing the number of discrete character states for continuous characters generates well-resolved trees that do not reflect phylogeny. Integr. 9(4): 531-41.
- Hoyal Cuthill JF. (2015). The morphological state space revisited: what do phylogenetic patterns in homoplasy tell us about the number of possible character states? Interface Focus. 5(6): 20150049.
- Hoyal Cuthill JF. (2015). The size of the character state space affects the occurrence and detection of homoplasy: modelling the probability of incompatibility for unordered phylogenetic characters. J Theor Biol. Feb(366):24-32.
- Mounier A, Balzeau A, Caparros M, Grimaud-Hervé D. (2016). Brain, calvarium, cladistics: A new approach to an old question, who are modern humans and Neandertals? J Hum Evol. Mar(92):22-36.
- Pagel MD and Harvey PH. (1988). Recent developments in the analysis of comparative data. Q Rev Biol. 63(4):413-40.
Cladistics
No comments:
Post a Comment