Intron Definition
An intron is a long stretch of noncoding DNA found between exons (or coding regions) in a gene. Genes that contain introns are known as discontinuous or split genes as the coding regions are not continuous. Introns are found only in eukaryotic organisms.
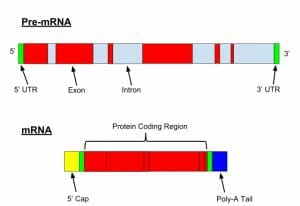
Here we see the structure of a pre-mRNA (or hrRNA) and a mature mRNA following mRNA processing (splicing, the addition of a 5′-cap and a poly-A tail).
Intron Discovery
Introns were discovered in 1977 with the introduction of DNA sequencing. While it was known that mature eukaryotic mRNA molecules were shorter than the initial transcripts, it was believed that the transcripts were simply trimmed at the ends. When the two molecule types were sequenced it was revealed that this was not the case; much of the removed transcript came from internal regions rather than the extreme ends. This prompted extensive research into how introns were removed from transcripts, and what their role might be.
Intron Structure
In general, introns are much longer than exons; they can make up as much as 90% of a gene and can be over 10,000 nucleotides long. Introns are prevalent in genes; over 90% of human genes contain introns with an average of nine introns per gene.
An intron is a stretch of DNA that begins and ends with a specific series of nucleotides. These sequences act as the boundary between introns and exons and are known as splice sites. The recognition of the boundary between coding and non-coding DNA is crucial for the creation of functioning genes. In humans and most other vertebrates introns begin with 5′ GUA and end in CAG 3′. There are other conserved sequences found in introns of both vertebrates and invertebrates including a branch point involved in lariat (loop) formation.
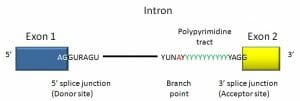
Here we see a consensus sequence for a vertebrate intron. The intron begins with GUR and ends in a polypyrimidine tract followed by YAG.
Intron Function
While introns were initially – and to an extent still are – considered “junk DNA”, it has been shown that introns likely play an important role in regulation and gene expression. As introns cause an increase in gene length, this increases the likelihood of crossing over and recombination between sister chromosomes. This increases genetic variation and can result in new gene variants through duplications, deletions, and exon shuffling. Introns also allow for alternative splicing. This allows a single gene to encode multiple proteins as the exons can be assembled in multiple ways.
Splicing
During transcription RNA polymerase copies the entire gene, both introns and exons, into the initial mRNA transcript known as pre-mRNA or heterogeneous nuclear RNA (hrRNA). As introns are not transcribed, they must then be removed before translation can occur. The excision of introns and the connection of exons into a mature mRNA molecule occurs in the nucleus and is known as splicing.
Introns contain a number of sequences that are involved in splicing including spliceosome recognition sites. These sites allow the spliceosome to recognise the boundary between the introns and exons. The sites themselves are recognised by small nucleolar ribonucleoproteins (snRNPs). There are a number of snRNPs involved in mRNA splicing which combined create a spliceosome.
Splicing occurs in three steps:
- Cleavage of the phosphodiester bond between the exon and the GU at the 5′ end of the intron. One snRNP (U1) contains a complementary sequence to the 5′ splice site and binds there to initiate splicing.
- Formation of a lariat or loop structure. The free 5′ end of the intron connects to a branch site, a conserved sequence near the 3′ end of the intron. A second snRNP (U2) binds to the branch site and attracts U1 to initiate the lariat. The lariat is then formed by a phosphodiester bond between the free 5′ G and an A at the branch site.
- Cleavage of the phosphodiester bond between the second exon and the 3′ AG of the intron.
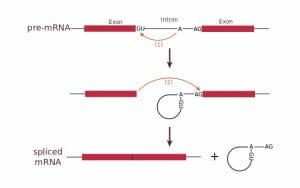
This figure shows the splicing of an intron through formation of a lariat. The intron is then removed leaving the two exons connected.
It is unknown how the snRNPs and the spliceosome identify which recognition sites to bind to given the that the introns can be thousands of base pairs long and there are many cryptic splice sites where the recognition sequences are found elsewhere in the gene. It is believed that certain proteins (for example, SR proteins), enhancers, and silencers are involved. Splicing silencers have also been implicated in human diseases.
Alternative splicing
Introns and the splicing mechanism also allow for alternative gene products in a process known as alternative splicing. Each discontinuous gene is made up of two or more exons, allowing for multiple ways in which the exons can be assembled. Alternative splicing can result in two to hundreds of different mRNAs. Alternative splicing is common in some species but rare in others; it is found in over 80% of human genes but there are only three known cases in Saccharomyces cerevisiae (yeast).
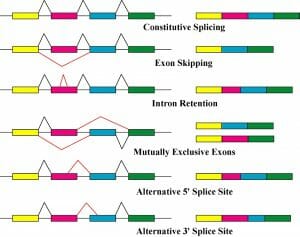
Alternative splicing can occur in a number of ways:
- Exon skipping: one (or more) exons are not included in the final mRNA
- Intron retention: part of the intron is not properly spliced and remains in the final mRNA
- Alternative splice site: the spliceosome removes part of one (or more) exon as well as the intron
rRNAs and tRNAs
Introns can also be found in both pre-rRNAs and pre-tRNAs. Introns in rRNAs are rare, with examples so far found only in lower eukaryotes. Unlike introns in other molecules, some rRNA introns have a unique characteristic – they are self-splicing. Self-splicing introns fall into a category known as Group I introns. Rather than relying on an external enzyme to perform the excision the introns themselves act as an enzyme known as a ribozyme. Ribozymes were discovered in the ciliate Tetrahymena in 1982 and revolutionized the way scientists viewed enzymes.
Introns in tRNAs are more common than those in rRNAs but much less prevalent than in mRNAs, particularly in vertebrates (i.e., 6% of human tRNAs). Introns in tRNAs are relatively short, ranging from 14 to 60 base pairs in length. The introns form part of the stem and loop structure of the tRNA, binding to a section of the anticodon arm. Removal of pre-tRNA introns is done by a single endonuclease.
Quiz
1. Which organisms do not have introns?
A. bacteria
B. fungi
C. protozoa
D. plants
2. Where does splicing occur?
A. cytosol
B. ribosomes
C. nucleus
D. chloroplasts
3. What molecules contain introns?
A. pre-mRNA
B. pre-rRNA
C. pre-tRNA
D. all of the above
References
- Brown, T. (2012).Introduction to genetics: a molecular approach Chs. 3, 5, and 6. New York, NY: Garland Science, Taylor & Francis Group, LLC. ISBN: 978-0-8153-6509-9.
- Wong, G. K.-S., Passey, D. A., Huang, Y.-Z., Yang, Z., & Yu, J. (2000). “Is “junk” DNA mostly intron DNA?”. Genome Research 10: 1672-1678.
- Wong, J. J.-L., Au, A. Y. M., Ritchie, W., & Rasko, J. E. J. (2015). Intron retention in mRNA: no longer nonsense. Bioessays 38: 41-49.
Intron
No comments:
Post a Comment