DNA Sequencing Definition
DNA sequencing is the process of determining the exact sequence of nucleotides within a DNA molecule. This means that by sequencing a stretch of DNA, it will be possible to know the order in which the four nucleotide bases – adenine, guanine, cytosine and thymine – occur within that nucleic acid molecule.
The necessity of DNA sequencing was first made obvious by Francis Crick’s theory that the sequence of nucleotides within a DNA molecule directly influenced the amino acid sequences of proteins. At the time, the belief was that a completely sequenced genome would lead to a quantum leap in understanding the biochemistry of cells and organisms.
History of DNA Sequencing
Watson and Crick’s discovery of DNA structure created the theoretical framework for understanding DNA replication and transcription. However, for nearly two decades, it was not possible to know the exact sequence of nucleotides within a DNA molecule. While proteins could be sequenced by enzymatic digestion, nucleotides were very similar to each other in their chemical composition, complicating the biochemical analysis of DNA.
The first DNA fragment to be sequenced belonged to a small virus called T4 bacteriophage that specifically infects Escherichia coli bacteria. An important gene in this organism codes for the enzyme, lysozyme. The amino acid sequence of this enzyme had been elucidated earlier through sequential digestion with the enzyme trypsin, and the DNA sequence was identified later. The crux of this method was to use a primer and a DNA polymerase enzyme to bind to specific sections of the DNA and to elongate the molecule in a manner that mimicked DNA polymerization in vivo.
In the mid-1970s, Frederick Sanger improved this initial method by using a plus-minus system for running a sequencing reaction. In this modified method, DNA polymerization initially occurred using radiolabeled nucleotides. After that, a short two-second pulse of polymerization was done by adding or omitting a single nucleotide in each reaction mixture. This created a set of eight reactions to give a definitive picture of the nucleotide sequence within a DNA molecule. The completed reactions were run on a polyacrylamide gel for analysis. In this manner, the first complete genome was sequenced, that of bacteriophage ϕX174.
Maxam and Gilbert further modified this method y busing radiolabeled DNA and chemicals (such as hydrazine) that would selectively induce the DNA molecule to break at certain bases. Once again, the results of this chemical digestion were analyzed on polyacrylamide gels.
One of the biggest breakthroughs in this field was the development of chain-termination technology using modified nucleotides by the Sanger lab. These modified nucleotides, were variants of deoxyribonucleotides (dNTPs) and were called dideoxynucleotides (ddNTPs). They had two missing oxygen atoms, at the 2’ and 3’ positions of the sugar molecule. The addition of a ddNTP made DNA polymerization stop, since the modified nucleotide could not form a covalent bond with an incoming dNTP. The accuracy, ease and robustness of this method, called the dideoxytermination method or Sanger sequencing, made it the definitive method for DNA sequencing in the subsequent years.
In 1983, polymerase chain reaction (PCR) for amplifying stretches of DNA was discovered. This had a big impact on DNA sequencing efforts since PCR could generate the high concentrations of DNA needed to run large sequencing reactions. In 1984, the complete genome of the Epstein Barr virus was elucidated, and over the next few years, the genomes of a number of organisms were sequenced. These included fungi such as Mycoplasma capricolum and S. cerevisiae, bacteria like E. coli, and invertebrates such as the roundworm C. elegans. In 1990, the human genome project was launched, and over the next decade, two groups worked towards completely sequencing the human genome. In 2001, the results of these efforts were published, having cost nearly 3 billion dollars, and starting a new era of human genomics. By that time, the genomes of nearly 600 viruses and viroids, a number of organelles and plasmids, bacteria, fungi, two animals and one plant had also been completely sequenced.
By the start of the 21st Century, a number of new methods were being developed for large-scale sequencing. These targeted entire chromosomes or large numbers of short DNA fragments. The development of these high-throughput sequencing technologies vastly reduced the time and cost involved in sequencing a genome. Today, even personal genomics is affordable and accessible, where an individual can obtain an accurate copy of her entire genome.
DNA Sequencing Methods
There are two main types of DNA sequencing. The older, classical chain termination method is also called the Sanger method. Newer methods that can process a large number of DNA molecules quickly are collectively called High-Throughput Sequencing (HTS) techniques or Next-Generation Sequencing (NGS) methods.
Sanger Method
The Sanger method relied on a primer that would bind to a denatured DNA molecule and initiate the synthesis of a single-stranded polynucleotide in the presence of DNA polymerase enzyme, using the denatured DNA as a template. In most circumstances, the enzyme would catalyze the addition of a nucleotide. A covalent bond would therefore form between the 3′ carbon atom of the deoxyribose sugar molecule in one nucleotide and the 5′ carbon atom of the next.
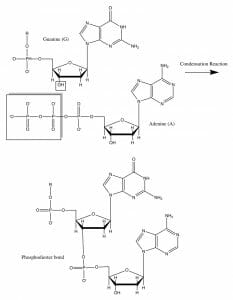
The image shows the formation of a phosphodiester covalent bond between a guanine and adenine nucleotide, through a condensation reaction. On closer examination, it becomes clear that the reaction occurs between the hydroxyl group on the 3’ carbon atom on a deoxyribose sugar and the phosphate moieties in the incoming nucleotide.
A sequencing reaction mixture, however, would have a small proportion of modified nucleotides that cannot form this covalent bond due to the absence of a reactive hydroxyl group, giving rise to the term ‘dideoxyribonucleotides’, i.e., they do not have a 2’ or 3’ oxygen atom when compared to the corresponding ribonucleotide. This would terminate the DNA polymerization reaction prematurely. At the end of multiple rounds of such polymerizations, a mixture of molecules of varying lengths, would be created.
In the earliest attempts at using the Sanger method, the DNA molecule was first amplified using a labeled primer and then split into four test tubes, each having only one type of ddNTP. That is, each reaction mixture would have only one type of modified nucleotide that could cause chain termination. After the four reactions were completed, the mixture of DNA molecules created by chain termination would undergo electrophoresis on a polyacrylamide gel, and get separated according to their length.
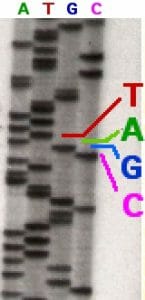
In the image above, a sequencing reaction with ddATP was electrophoresed through the first column. Each line represents a DNA molecule of a particular length, the result of a polymerization reaction that was terminated by the addition of a ddATP nucleotide. The second, third and fourth columns contained ddTTP, ddGTP, and ddCTP respectively.
With time, this method was modified so that each ddNTP had a different fluorescent label. The primer was no longer the source of the radiolabel or fluorescent tag. Also known as dye-terminator sequencing, this method used four dyes with non-overlapping emission spectra, one for each ddNTP.
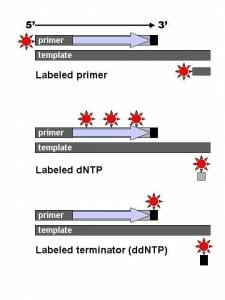
The image shows the difference between labeled primers, labeled dNTPs and dyed terminator NTPs.
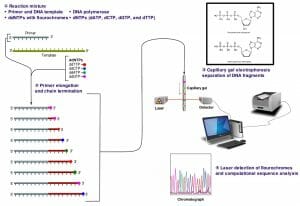
The image above shows a schematic representation of dye-terminator sequencing. There is a single reaction mixture carrying all the elements needed for DNA elongation. The reaction mixture also contains small concentrations of four ddNTPs, each with a different fluorescent tag. The completed reaction is run on a capillary gel. The results are obtained through an analysis of the emission spectra from each DNA band on the gel. A software program then analyzes the spectra and presents the sequence of the DNA molecule.
High Throughput Sequencing
Sanger sequencing continues to be useful for determining the sequences of relatively long stretches of DNA, especially at low volumes. However, it can become expensive and laborious when a large number of molecules need to be sequenced quickly. Ironically, though the traditional dye-terminator method is useful when the DNA molecule is longer, high-throughput methods have become more widely used, especially when entire genomes need to be sequenced. The human genome project cost nearly 3 billion dollars. In 2004, a large sum of money was pumped into the development of low-cost high-throughput sequencing technology, to potentially allow an entire human genome to be sequenced for less than 1000 dollars.
There were three major changes to the Sanger method. The first was the development of a cell free system for cloning DNA fragments. Traditionally, the stretch of DNA that needed to be sequenced was first cloned into a prokaryotic plasmid, and amplified within bacteria before being extracted and purified. High throughput sequencing or next-generation sequencing technologies no longer relied on this labor-intensive and time-intensive procedure. Secondly, these methods created space to run millions of sequencing reactions in parallel. This was a huge step forward from the initial methods where eight different reaction mixtures were needed to produce a single reliable nucleotide sequence. Finally, there is no separation between the elongation and detection steps. The bases are identified as the sequencing reaction proceeds. While HTS decreased cost and time, their ‘reads’ were relatively short. That is, in order to assemble an entire genome, intensive computation was necessary, that put together millions of short stretches of sequenced DNA to create the overall nucleotide sequence of a chromosome or genome.
The advent of HTS has vastly expanded the applications for genomics. DNA sequencing has now become an integral part of basic science, translational research, medical diagnostics, and forensics.
Uses of DNA Sequencing
Traditional, chain-termination technology and HTS methods are used for different applications today. Sanger sequencing is now used mostly for de novo initial sequencing of a DNA molecule to obtain the primary sequence data for an organism or gene. The relatively short ‘reads’ coming off a HTS reaction (30-400 base pairs compared to the nearly a thousand base pair ‘reads’ from Sanger sequencing methods) make it difficult to create the entire genome of any organism from HTS methods alone. Occasionally, Sanger sequencing is also needed to validate the results of HTS.
On the other hand, HTS allows the use of DNA sequencing to understand single-nucleotide polymorphisms – among the most common types of genetic variation within a population. This becomes important in evolutionary biology as well as in the detection of mutated genes that can result in disease. For instance, sequence variations in samples from lung adenocarcinoma allowed the detection of rare mutations associated with the disease. The chromatin binding sites for specific nuclear proteins can also be accurately identified using these methods
Overall, DNA sequencing is becoming an integral part of many different applications.
Diagnostics
Genome sequencing is particularly useful for identifying the causes in rare genetic disorders. While more than 7800 diseases are associated with a Mendelian inheritance pattern, less than 4000 of those diseases have been definitively linked to a specific gene or mutation. Early analysis of the exon-genome, or exome, consisting of all the expressed genes of an organism, showed promise in identifying the causal alleles for many inherited illnesses. In one particular case, sequencing the genome of a child suffering from a severe form of inflammatory bowel disease connected the illness to a mutation in a gene associated with inflammation – XIAP. While the patient initially showed multiple symptoms suggestive of an immune deficiency, a bone marrow transplant was recommended based on the results of DNA sequencing. The child subsequently recovered from the ailment.
In addition, HTS has been an important player in developing a greater understanding of tumors and cancers. Understanding the genetic basis of a tumor or cancer enables doctors to have an extra tool in their kit for making diagnostic decisions. The Cancer Genome Atlas and International Cancer Genome Consortium have sequenced a large number of tumors and demonstrated that these growths can vary vastly in terms of their mutational landscape. This has also given a better understanding of the kind of treatment options that are ideal for each patient. For instance, the sequencing of the breast cancer genome identified two genes – BRCA1 and BRCA2 – whose pathogenic variants have an enormous impact on the likelihood of developing breast cancer. People with some pathogenic alleles even choose to have preventive surgeries such as double mastectomies.
Molecular Biology
DNA sequencing is now an integral part of most biological laboratories. It is used to verify the results of cloning exercises to understand the effect of particular genes. HTS technologies are used to study variations in the genetic compositions of plasmids, bacteria, yeast, nematodes or even mammals used in laboratory experiments. For instance, a cell line derived from breast cancer tissue, called HeLa, is used in many laboratories around the world and was earlier considered as a reliable cell line representing human breast tissue. Recent sequencing results have demonstrated large variations in the genome of HeLa cells from different sources, thereby reducing their utility in cell and molecular biology.
DNA sequencing gives insight into the regulatory elements within the genome of every cell, and the variations in their activity in different cell types and individuals. For instance, a particular gene may be permanently turned off in some tissues, while being constitutively expressed in others. Similarly, those with susceptibility for a specific ailment may regulate a gene differently from those who are immune. These differences in the regulatory regions of DNA can be demonstrated through sequencing and can give insight into the basis for a phenotype.
Recent advances have even allowed individual laboratories to study structural variations in the human genome – an undertaking that needed global collaboration two decades ago.
Forensics
The ability to use low concentrations of DNA to obtain reliable sequencing reads has been extremely useful to the forensic scientist. In particular, the potential to sequence every DNA within a sample is attractive, especially since a crime scene often contains genetic material from multiple people. HTS is slowly being adopted in many forensics labs for human identification. In addition, recent advances allow forensic scientists to sequence the exome of a person after death, especially to determine the cause of death. For instance, death due to poisoning will show changes to the exome in affected organs. On the other hand, DNA sequencing can also determine that the deceased had a preexisting genetic ailment or predisposition. The challenges in this field include the development of extremely reliable analysis software, especially since the results of HTS cannot be manually examined.
Quiz
1. Why did DNA sequencing develop more slowly than protein sequencing?
A. The structure of DNA was discovered much after protein structures were determined
B. The development of ddNTPs took a lot of time
C. Restriction enzymes for splicing DNA were not available
D. Compared to amino acids, nucleotides are more similar to each other chemically
2. Approximately, how much did it cost to initially sequence a human genome?
A. 1 million dollars
B. 100 million dollars
C. 300 million dollars
D. 3 billion dollars
3. Which organism was the first to have a completely sequenced genome?
A. A bacteriophage – a virus that infects bacteria
B. A roundworm – an invertebrate that is often used in genetics laboratories
C. A fungus – a yeast that is has been used to make bread and wine for millennia
D. A bacteria – a prokaryote commonly found in the intestines of humans
References
- Børsting, C. and Morling, N., (2015) “Next generation sequencing and its applications in forensic genetics” Forensic Science International: Genetics 18: 78-89
- Fox, S., Filichkin, S., Mockler, T. C., (2009) “Applications of ultra-high-throughput sequencing” Methods Mol Biol. 553:79-108
- Hall N., (2007). “Advanced sequencing technologies and their wider impact in microbiology”J. Exp Biol.210:1518-1525
- Heather J. M., Chain, B., (2016) “The sequence of sequencers: The history of sequencing DNA” Genomics 107(1):1-8
- Lander et al., (2001) “Initial sequencing and analysis of the human genome” Nature 409: 860-921
- Reuter J. A., Spacek D., and Snyder M. P., (2015) “High-Throughput Sequencing Technologies” Mol. Cell. 58(4): 586-597
- van Dijk, E. L. et al., (2014) “Ten years of next-generation sequencing technology” Trends in Genetics 30(9): 418-426
DNA Sequencing
No comments:
Post a Comment